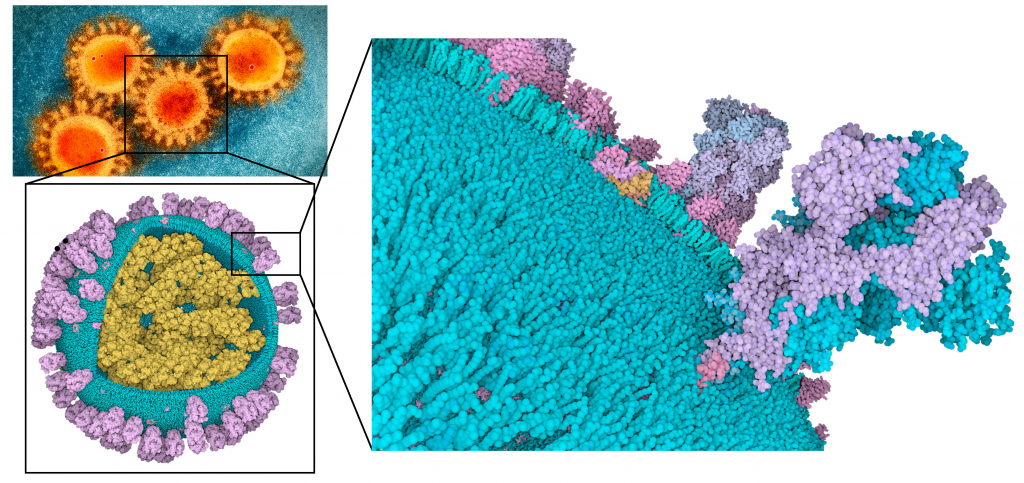
OpenPandemics: COVID-19
In collaboration with IBM, we are joining the World Community Grid to dock millions of small molecules to different proteins that play a role in the SARS-CoV2 life cycle. Promising molecules will be experimentally tested by our collaborators at Scripps, and hopefully evolve to become COVID-19 antivirals.
For that, we are going to screen conventional binders as well as covalent (i.e. irreversible) inhibitors. Using simple chemical reactions, many commercially available molecules can be modified to carry a reactive group. We identified tens of millions of such molecules in the ZINC database. Now, we are leveraging our Reactive Docking protocol to find molecules that can covalently attach to the protein. The new active molecules will provide not only new tools to fight the infection, but also molecular probes to deepen our understanding of the many obscure and still unknown aspects of the virus biology.
Thanks to the World Community Grid infrastructure and to volunteers who donate the computing power of their workstations, laptops, cell phones, and other devices, we will dock a very large number of molecules, possibly the largest covalent screening ever performed.
For more information on the collaborating labs, News, FAQ and statistics, check the OpenPandemics: COVID-19 Project page.
The COVID-NineTeam
The Team is subdivided in two sub-teams: Structure Team and Computing Team.
 The Structure Team curates and validates the target protein structures used for the dockings. They analyze and prepare the structures looking for potential binding pockets as well as residues suitable to be targeted by the covalent inhibitors. Their role is to guarantee the quality of the target structures and that they capture their conformational variability in order to guarantee the success of the dockings. The Structure Team is working in collaboration with Martina Maritan from the Goodsell Lab to build mesoscale models of the full virus. The full viral model images have made built by Ludovic Autin.
The Structure Team curates and validates the target protein structures used for the dockings. They analyze and prepare the structures looking for potential binding pockets as well as residues suitable to be targeted by the covalent inhibitors. Their role is to guarantee the quality of the target structures and that they capture their conformational variability in order to guarantee the success of the dockings. The Structure Team is working in collaboration with Martina Maritan from the Goodsell Lab to build mesoscale models of the full virus. The full viral model images have made built by Ludovic Autin.
 The Computing Team is responsible of the preparation of the ligands to be docked, preparing the structure of commercially available compounds, and performing the in silico enumerations that generate new libraries of synthetically accessible derivatives, thereby expanding the sampling of the chemical space. They are also managing the hardware and software infrastructure for managing both input and output data, performing the automated analyses steps on the results, and storing them in the database.
The Computing Team is responsible of the preparation of the ligands to be docked, preparing the structure of commercially available compounds, and performing the in silico enumerations that generate new libraries of synthetically accessible derivatives, thereby expanding the sampling of the chemical space. They are also managing the hardware and software infrastructure for managing both input and output data, performing the automated analyses steps on the results, and storing them in the database.
For more details about the people involved, check out the Lab Members page.
…So when there will be a new drug?
Our effort, as well as many others that are going on right now, aims at the generation of new data that will be essential to improve our understanding of the biological processes of the SARS-CoV2 virus. With the help of the WCG volunteers and our scientific collaborators, we hope to identify new chemical compounds that can interfere with these processes, and provide the foundations for the design of effective drugs. This initial discovery phase is very difficult because of all the unknowns we have to face with such a new threat, and our job will be essential to make it as short as possible.
Nevertheless, no matter how good our results will be, they are just the beginning of the long journey that will eventually lead to a drug, which takes years and many more efforts before it can reach the bedside.