Our Research
Using computational tools to uncover the rules of biology and enable new therapeutic discoveries
Developing Computational Methods
We develop new, cutting-edge computational methods to predict and analyze the interactions of small organic molecules (either synthetic or from natural sources) with biological macromolecular structures such as proteins and nucleic acids.
Understanding Biological Mechanisms
By combining these tools with other molecular modeling techniques, we investigate biological processes from mechanistic and thermodynamic perspectives. This work helps us unravel the physico-chemical principles that govern complex biological events.
Applications to Drug Discovery
We apply our methods to rapidly explore chemical space and identify molecular modulators of therapeutically relevant targets. These efforts are carried out in collaboration with researchers at Scripps Research, as well as partners in academia and pharmaceutical industry.
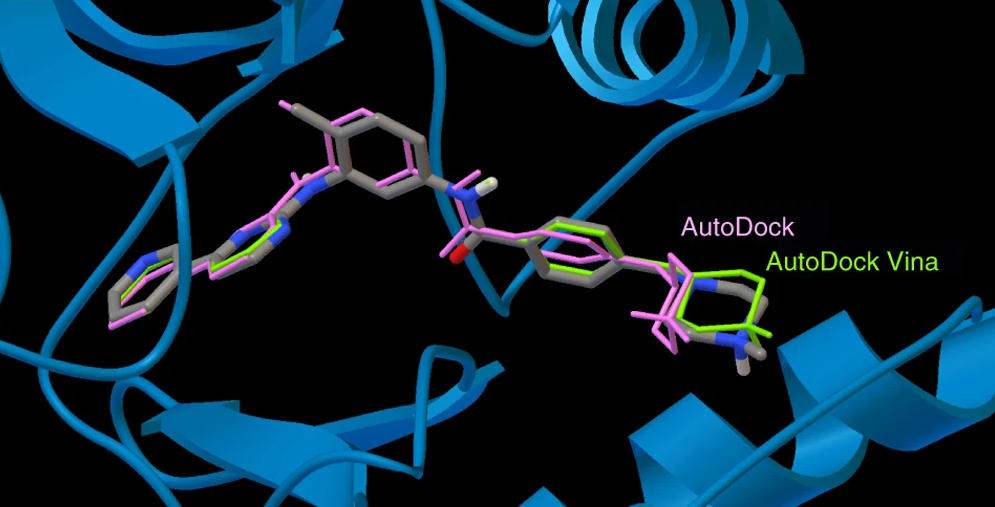
AutoDock suite
We develop computational tools for structure-based drug design, with a focus on molecular docking. AutoDock and AutoDock Vina, originally created in the Molecular Graphics Lab (now CCSB), are among the most widely used open-source docking programs. Building on this foundation, we are developing next-generation docking engines and virtual screening tools with improved energy potentials, more accurate binding free energy estimates, and innovative protocols for drug design.
Our goal is to make docking faster, more accurate, and more chemically meaningful. To achieve this, we are improving the description of chemical, physical, and thermodynamic terms, including torsional preferences of rotatable bonds, non-covalent interactions, and solvation effects, drawing on modern force fields, electronic structure methods, and molecular dynamics simulations. In parallel, we explore new search algorithms and leverage GPU acceleration to navigate the energy landscape efficiently and to scale virtual screening.
Relevant publications
Industry partners



Relevant publications
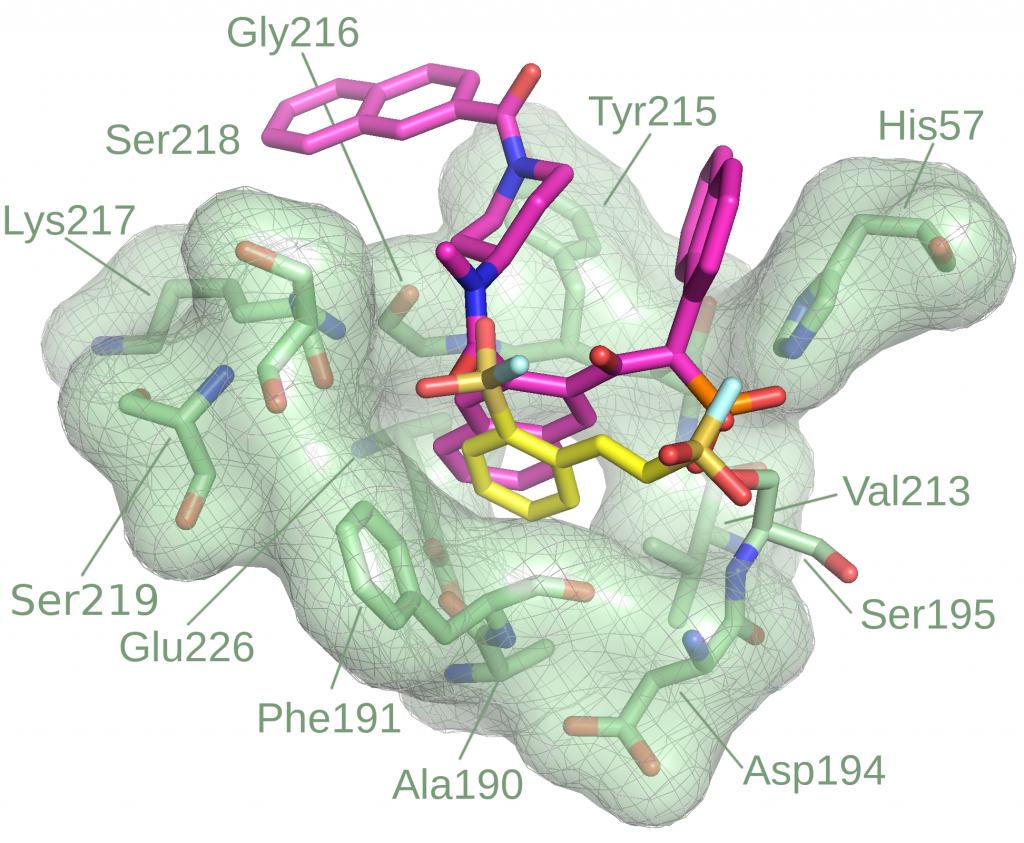
Covalent inhibitor high-throughput screening
Modeling of covalent binders is non-trivial, especially in high-throughput settings. While several covalent docking methods exist, most are effective only when the target structure and covalent binding site are already known. To overcome this barrier, we are developing Reactive Docking, a method that predicts covalent binding sites by simulating the reaction between a protein residue and a ligand warhead. This approach supports diverse residues and warhead chemistries. It has been successfully applied to multiple targets and is suitable for HTVS to discover novel covalent binding sites across different proteins.
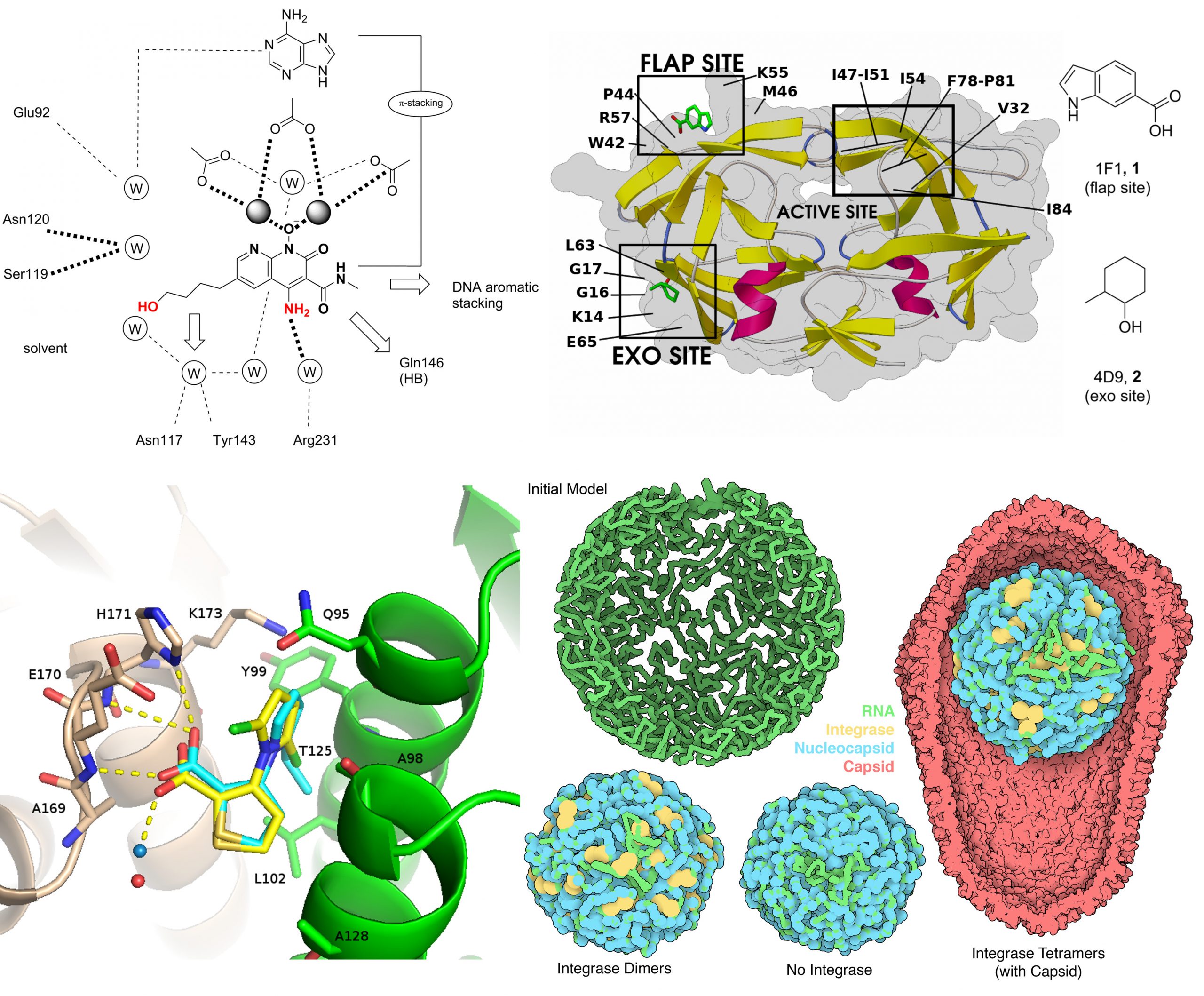
HIV-1 life cycle
The search for drugs that target different stages of the HIV-1 life cycle remains a subject of great interest, since current therapies cannot fully eradicate the infection. We are focusing on the HIV-1 capsid protein, using a strategy that combines conventional and reactive docking for the virtual screening of small molecules that could modulate the viral assembly. In particular, our Reactive Docking approach is applied to identify small molecules with SuFEx warheads that could tightly bind to the identified pocket.
Relevant publications
Initiative
Relevant publications
Molecular modeling applications
We use molecular modeling techniques such as molecular dynamics, pharmacophore modeling, and virtual screenings to study protein-ligand and protein-protein interactions. These approaches help reveal the structural basis of drug action, immune regulation, RNA processing, and other biological processes, guiding therapeutic discovery and design.